5 Introduction to R part II: Visualisation
If you are currently participating in a timetabled BIOS103 QS workshop, please ensure that you cover all of this section’s content and complete this week’s summative assessment in the BIOS103 Canvas Course. There is no formative assessment this week.
Our brains are hardwired to recognise patterns, which means we can understand information much more easily when it’s presented visually rather than as raw numbers. In today’s data-driven world, the ability to visualise information effectively is more important than ever. Learning to visualise data will equip you with essential skills for analysing and interpreting information, allowing you to communicate insights clearly and persuasively.
Effective visualisation also fosters critical thinking. It helps you identify trends, relationships, and anomalies within datasets, enhancing your analytical abilities and preparing you for future roles in research, industry, and beyond, where data-driven decision-making is key.
This week, we’re going to explore visualisation in R, focusing on creating impactful graphics using its native capabilities. Our objectives include:
- Histograms: Visualising the distribution of continuous variables.
- Boxplots: Showing data spread between groups and highlighting outliers.
- Scatterplots: Displaying relationships between two continuous variables.
By the end of this chapter, you will be equipped to create these essential visualisations, enhancing your analytical skills and bringing your data to life!
In an effort to ensure that this week’s chapter is relevant to your microbiology lab practicals, I have carefully crafted the following scenario to provide a bit more context for our visualisation.
5.1 Mushroom Compost Scenario
MegaMush, a mushroom compost company based in the Netherlands, is trying to identify an issue with its pre-pasteurisation composting process across its five operational production sites.
You have been hired as an independent bioscience data consultant and have been tasked with summarising and visualising a dataset compiled over a period of 1 year and consolidated from each location.
Specifically, you have been asked to generate the following for incorporation into a report that will be presented by the chief technical officer of the company to the extended board of directors:
- A summary table that shows the mean and standard deviations of the composting temperature, moisture, and viable bacterial count, VBC (cfu/g), at each site.
- A histogram showing the distribution of estimated viable bacterial count, VBC from across all samples.
- A grouped boxplot showing the distributions of estimated VBC for each site.
- A scatterplot that shows the relationship between temperature and VBC.
- Any additional visualisations (histogram, boxplot or scatterplot) that identify other interesting features in the data.
5.1.1 The dataset
You can download the raw data here or from the backup link.
5.1.2 Further information
The compost temperature and moisture content were measured directly during the sampling process using calibrated thermometers and moisture probes.
After collection, samples were immediately placed into sterile containers to prevent cross-contamination and transported to the central laboratory under refrigerated conditions at 4°C. To ensure the integrity of microbial counts, all samples were processed within 24 hours of collection.
In the laboratory, 0.5g of each compost sample was dissolved in 10mL of sterile reagent and mixed thoroughly to create the starting solution for subsequent serial dilution. In each case, a 1/1000 serial dilution was performed and 0.1mL of the diluted sample was plated onto selective agar media. Plates were incubated for 48 hrs at 30°C before the number of viable colonies were counted.
This procedure was performed regularly for each site, ensuring consistent data collection on microbial counts, temperature, and moisture content across all locations over the course of a year.
Your tables and figures need to be formatted according to the MegaMush report template
5.2 Inspecting and Summarising
Right then, let’s start. You know the drill by now:
- Create a New R Project
- Download the dataset and move to project folder
- Create a new script file and save it with a sensible filename
- Read your dataset to a new variable called
data
That should create a data variable in my environment window. I could click on it and look at the data, but that method was so “last week”. This time I’m going to simply add the line head(data) to my script and run it to spit out the first 5 rows of my data (including headers) in my console. Here’s what my script (and its output) looks like so far:
# Script File: mushroom_summary.R
# Read the data
data <- read.csv("compost_999.csv")
# Output the first 5 rows in console
head(data)## SampleID Location DateTime DayOfWeek Temperature Moisture
## 1 MAA_20230802 Maastricht 2023-08-02 Wednesday 54.8 71.8
## 2 ROT_20230611 Rotterdam 2023-06-11 Sunday 55.5 61.8
## 3 UTR_20230818 Utrecht 2023-08-18 Friday 68.2 78.5
## 4 AMS_20230913 Amsterdam 2023-09-13 Wednesday 57.6 73.0
## 5 UTR_20230619 Utrecht 2023-06-19 Monday 70.5 76.3
## 6 GRO_20231109 Groningen 2023-11-09 Thursday 52.9 70.3
## Viable.counts
## 1 153
## 2 144
## 3 139
## 4 149
## 5 133
## 6 155OK. So it doesn’t look too pretty in my console, but I can glean the important information - i.e. the data headers and the types of data that are in each column. The headers are self explanatory , except perhaps for Viable.counts which is a direct count of the number of observed colonies formed on each sample plate. This is not to be confused with Viable Bacterial Count, VBC, defined as the number of colony forming units per gram (cfu/g).
Calculating VBC from Viable Counts
To calculate the Viable Bacterial Count (VBC) from the Viable Counts observed on the agar plates, we need to consider the specific parameters of the laboratory procedure. Here’s how the calculation works step-by-step:
- Viable Counts: The number of colonies (cfu) observed on an agar plate after incubation. Let’s denote this VC
- Dilution Factor: Remember, all starting solutions have undergone a 1/1000 serial dilution, we must first multiply by a factor of 1000.
- Volume Plated: From each diluted sample, 0.1 mL was plated onto the agar. This means that only 0.1mL of the diluted solution contributes the visible colonies counted. To express the counts in terms of 1mL, we must multiply by another factor of 10.
- Volume of Starting Solution: At this points we have units of cfu/mL. To figure out how many cfu there were in the starting solution of 10 mL we need to multiply by yet another factor of 10.
- Weight of Sample: Finally, since 0.5 g of each compost sample was used in the initial preparation, we need to express the results per gram. To convert out counts to reflect the weight of the sample, we divide by the weight used, which is 0.5 g. This can be expressed as multiplying by a factor of 2.
Putting all this together, the equation to calculate VBC from the observed Viable Counts can be expressed as:
\[ \text{VBC (cfu/g)} = \frac{\text{VC} \times (dilution\ factor)}{(spread\ plated\ volume)} \times \frac{(starting\ solution\ volume)}{(sample\ mass)} \]
\[ \text{VBC (cfu/g)} = \frac{\text{VC} \times (1000)}{(0.1)} \times \frac{(10)}{(0.5)} \]
Thus, the final calculation simplifies to:
\[ \text{VBC (cfu/g)} = \text{VC} \times 1000 \times 10 \times 10 \times 2 = \text{VC} \times 200,000 \]
This equation indicates that for every viable colony observed, the equivalent VBC is determined by multiplying by a factor of 200,000 to account for the dilution, volume plated, and sample weight, providing a standardised measure of viable bacteria per gram of compost.
5.2.1 Summary Table
Let’s tick the first task from the scenario off our list of things to do. Let’s create a summary table that shows the mean and standard deviations of the composting temperature, moisture, and viable bacterial count, VBC (cfu/g), at each site.
Curses! I’ve completely forgotten how to create a summary table in R. It’s the end of the world!
No it’s not. Let’s do the obvious (and easy) thing and revisit the script we used in the last chapter to generate a summary table and see if we can tweak things to make it work for us again.
Here’s the original script (or the first couple of lines at least).
# Read and filter data
data <- read.csv("pantheria_999.csv")
data_filtered <- data[data$NeonateBodyMass_g != -999, ]
# Calculate summary statistics for each Family
summary_table <- aggregate(NeonateBodyMass_g ~ Family, data_filtered, function(x)
c(Mean = mean(x), Median = median(x), Max = max(x), Min = min(x), SD = sd(x), Count = length(x)))
# Convert the list columns to individual columns
summary_table <- do.call(data.frame, summary_table)To adapt this script to work for my new “compost_999.csv” I need to consider the following things:
- Update the dataset: I need to update my
read.csv()function by replacingpantheria_999.csvwithcompost_999.csv. - Filter step removal: I know that my compost_999.csv dataset has no missing or erroneous data (please take my word for this) so I don’t need to worry about creating a new
data_filteredvariable. I can delete the line and update the rest of the code to just use my initialdatavariable throughout. - Adjust the grouping and variables: In my aggregate function I no longer need to summarise the
NeonateBodyMass_gbyFamily. Instead I want to summariseTemperature,Moisture, andViable.countsbyLocation. - Simplify descriptive statistics: I only need to calculate the mean and standard deviation within my aggregate function this time (forget about median, min and max).
With all that in mind, here’s how I’d update the script:
data <- read.csv("compost_999.csv")
# Calculate summary statistics for Temperature, Moisture, and Viable counts for each Location
summary_table <- aggregate(cbind(Temperature, Moisture, Viable.counts) ~ Location,
data,
function(x) c(Mean = mean(x), SD = sd(x)))
# Convert the list columns to individual columns
summary_table <- do.call(data.frame, summary_table)
# Output table in console
print(summary_table)## Location Temperature.Mean Temperature.SD Moisture.Mean Moisture.SD
## 1 Amsterdam 58.25165 2.126257 74.94286 2.028253
## 2 Groningen 54.67905 1.852581 70.03905 1.869157
## 3 Maastricht 56.84135 2.105592 72.42500 2.019793
## 4 Rotterdam 56.82762 2.015302 62.32762 1.971016
## 5 Utrecht 69.69368 2.051456 75.57263 1.743159
## Viable.counts.Mean Viable.counts.SD
## 1 150.5934 3.602401
## 2 152.0381 3.116099
## 3 150.7404 3.424626
## 4 142.6571 3.292849
## 5 134.0000 3.497720The biggest difference between the new and old scripts is the use of the cbind() function within my aggregate() function. The cbind() function allows me to group together multiple variables (Temperature, Moisure, Viable Counts) in order to calculate and present their descriptive stats simultaneously. Nice.
Our summary table is nearly complete. The last thing I need to do is calculate two more columns for the mean and standard deviations of the VBC (cfu/g). As I explained above, to get values of VBC I need to multiply my viable counts by a factor of 20,000. Here’s how I’d update my summary table script:
data <- read.csv("compost_999.csv")
# Calculate summary statistics for Temperature, Moisture, and Viable counts for each Location
summary_table <- aggregate(cbind(Temperature, Moisture, Viable.counts) ~ Location,
data,
function(x) c(Mean = mean(x), SD = sd(x)))
# Convert the list columns to individual columns
summary_table <- do.call(data.frame, summary_table)
# Define variable factor according to calculation for VBC. Divide by 1e7 to express answers in standard form.
factor <- 2e5 / 1e7
# Calculate additional columns vbc_mean and vbc_sd using factor
summary_table$vbc_mean <- summary_table$Viable.counts.Mean * factor
summary_table$vbc_sd <- summary_table$Viable.counts.SD * factor
# Save table to file
write.csv(summary_table, "compost_summary.csv", row.names = FALSE)
# Output table in console
print(summary_table)## Location Temperature.Mean Temperature.SD Moisture.Mean Moisture.SD
## 1 Amsterdam 58.25165 2.126257 74.94286 2.028253
## 2 Groningen 54.67905 1.852581 70.03905 1.869157
## 3 Maastricht 56.84135 2.105592 72.42500 2.019793
## 4 Rotterdam 56.82762 2.015302 62.32762 1.971016
## 5 Utrecht 69.69368 2.051456 75.57263 1.743159
## Viable.counts.Mean Viable.counts.SD vbc_mean vbc_sd
## 1 150.5934 3.602401 3.011868 0.07204801
## 2 152.0381 3.116099 3.040762 0.06232198
## 3 150.7404 3.424626 3.014808 0.06849252
## 4 142.6571 3.292849 2.853143 0.06585699
## 5 134.0000 3.497720 2.680000 0.06995439This script should work for you too. Update your existing script with the code above and check that you get the same output as that above for the example dataset.
Did you notice that I used the write.csv() in the penultimate line? This means that you should now see a file called summary_table.csv in your Files window. You should import this file into a new Excel workbook, format the numbers to a sensible precision, style it how you like, and then cut and paste it into your MegaMush report template. Don’t forget to include a table caption according to the unbreakable rules stated in Chatper 1.
5.3 Histogram
Let’s complete the second task on our list: Create a histogram showing the distribution of estimated viable bacterial count, VBC from across all samples.
Create a new script file, copy and paste the code below and run the whole file (click the Source button). You should see something like that in figure 5.1 in your Plots window.
# Your R code for generating the histogram
# Load the data
data <- read.csv("compost_999.csv")
# Calculate the conversion factor
factor <- 2e5 / 1e7
# Calculate VBC column
data$vbc <- data$Viable.counts * factor
# Calculate the mean VBC
vbc_mean <- mean(data$vbc)
# Generate a simple histogram for all "VBC"
hist(data$vbc,
breaks = 20,
main = "",
xlab = expression(VBC ~ "(" ~ x10^7 ~ cfu/g ~ ")"),
ylab = "Frequency",
col = "lightblue",
border = "black")
# Add a vertical line for the mean
abline(v = vbc_mean, col = "red", lwd = 2, lty = 2)
# I like a box around my figures
box()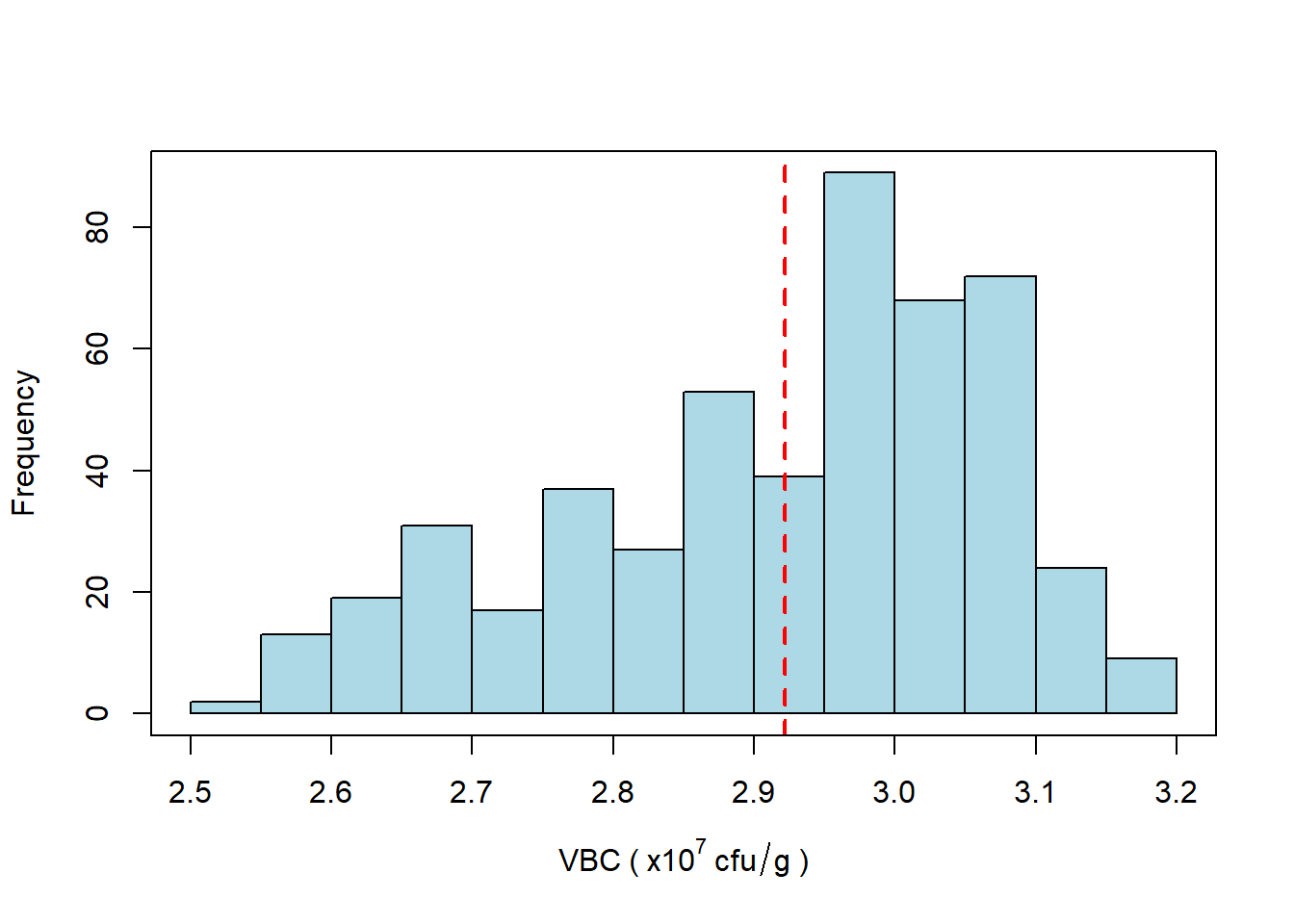
Figure 5.1: Histogram of VBC of samples taken from all sites over a period of 1 year. The mean VBC (dotted red line) is \(2.9 \times 10^7\) cfu/g.
Let’s break this code down comment by comment:
- Load the Data
- Purpose: This line uses the
read.csv()function to read in a CSV file named “compost_999.csv” and stores its content in a data frame variable calleddata
- Calculate the Conversion Factor
- Purpose: As for the summary table in the section above, this line calculates a conversion factor to convert viable counts into VBC in units of \(\times 10^7\) cfu/g.
- Scientific Notation:
2e5means \(2 \times 10^5\) and1e7means \(1 \times 10^7\). - Why divide by 1e7?: So that I can quote all VBC values in standard form, e.g. \(1.2 \times 10^7\) cfu/g.
- Calculate VBC column:
- Purpose: This line creates a new column
vbcin thedatadata frame. It calculates VBM by multiplying theViable.countscolumn by the conversion factor. - New Column:
data$vbcstores the calculated VBC for each row in the data frame.
- Calculate the mean VBC
- Purpose: This line computes the mean of the values in the
vbccolumn and stores it to a new variable calledvbc_mean. I’ll use this later shortly when I make my histrogram plot.
- Generate a Simple Histogram for all VBC
hist(data$vbc,
breaks = 20,
main = "",
xlab = expression(VBC ~ "(" ~ x10^7 ~ cfu/g ~ ")"),
ylab = "Frequency",
col = "lightblue",
border = "black")- Purpose: This block generates a histogram of the
vbcvalues. - Parameters:
breaks = 20: Specifies the number of bins (bars) in the histogram.main = "": Prevents a title from showing (I’ll use a caption later when I import the image to a document).xlab: Customises the x-label. Theexpression()function is used to format it, which displays the label as \(VBC (\times 10^7 cfu/g)\).ylab = Frequency: Sets the y-axis label to “Frequency”.col = "lightblue": Sets the color of the bars in the histogram to light blue.border = "black": Sets the border color of the bars to black
- Add a vertical line for the mean
- Purpose: This line adds a vertical dashed line to the histogram at the mean value
vbc_mean. - Parameters:
v = vbc_mean: Specifies the x-coordinate where the line is drawn (at the mean value).col = red: Sets the color of the line to red.lwd = 2: Sets the line width to 2 (give a thicker line than the default).lty = 2: Specifies the line type as dashed.
- I like a box around my histogram
- Purpose: This command make sure there is a full box around the histogram. By default, only the x and y axes will show otherwise.
How useful is this histogram?
Honestly? Not very. If you were hoping for a nice bell-curve (normal) distribution then you will be disappointed. All the histogram really tells us is that values of VBC are broadly distributed in the range \(2.5 - 3.2 \times 10^7\) cfu/g and that there seems to be a slight “skew” towards higher values in the range.
Oh well, at least you now know how to make a histogram using R in the future. Perhaps we could learn a bit more from our second type of visualisation: The Grouped Boxplot.
Feedback Please!
I really value your feedback on these materials for quantitative skills. Please rate them below and then leave some feedback. It’s completely anonymous and will help me improve things if necessary. Say what you like, I have a thick skin - but feel free to leave positive comments as well as negative ones. Thank you.
5.4 Grouped Boxplot
I love boxplots. There, I said it. They offer such a great way to visualise how your data is distributed, highlighting clear statistics like the median, quartiles, and potential outliers. But the real clincher is the their ability to compare multiple groups within your dataset at once to let you see if something interesting is going on between them.
Take a look at the script below. Start a new script, copy/paste the code and run all lines. You should see something similar to that shown in figure 5.2 appear.
# Your R code for generating a grouped boxplot.
# Load the data
data <- read.csv("compost_999.csv")
# Calculate the conversion factor
factor <- 2e5 / 1e7
# Calculate VBC column
data$vbc <- data$Viable.counts * factor
# Adjust the margins to prevent y-axis label from being cut off
par(mar = c(5, 5, 4, 2)) # Increase the second value for the left margin
# Generate a grouped boxplot for VBC and group by location
boxplot(vbc ~ Location,
data = data,
main = "",
xlab = "Location",
ylab = expression(VBC ~ "(" ~ x10^7 ~ cfu/g ~ ")"),
col = "lightblue",
border = "black",
cex.axis = 0.8)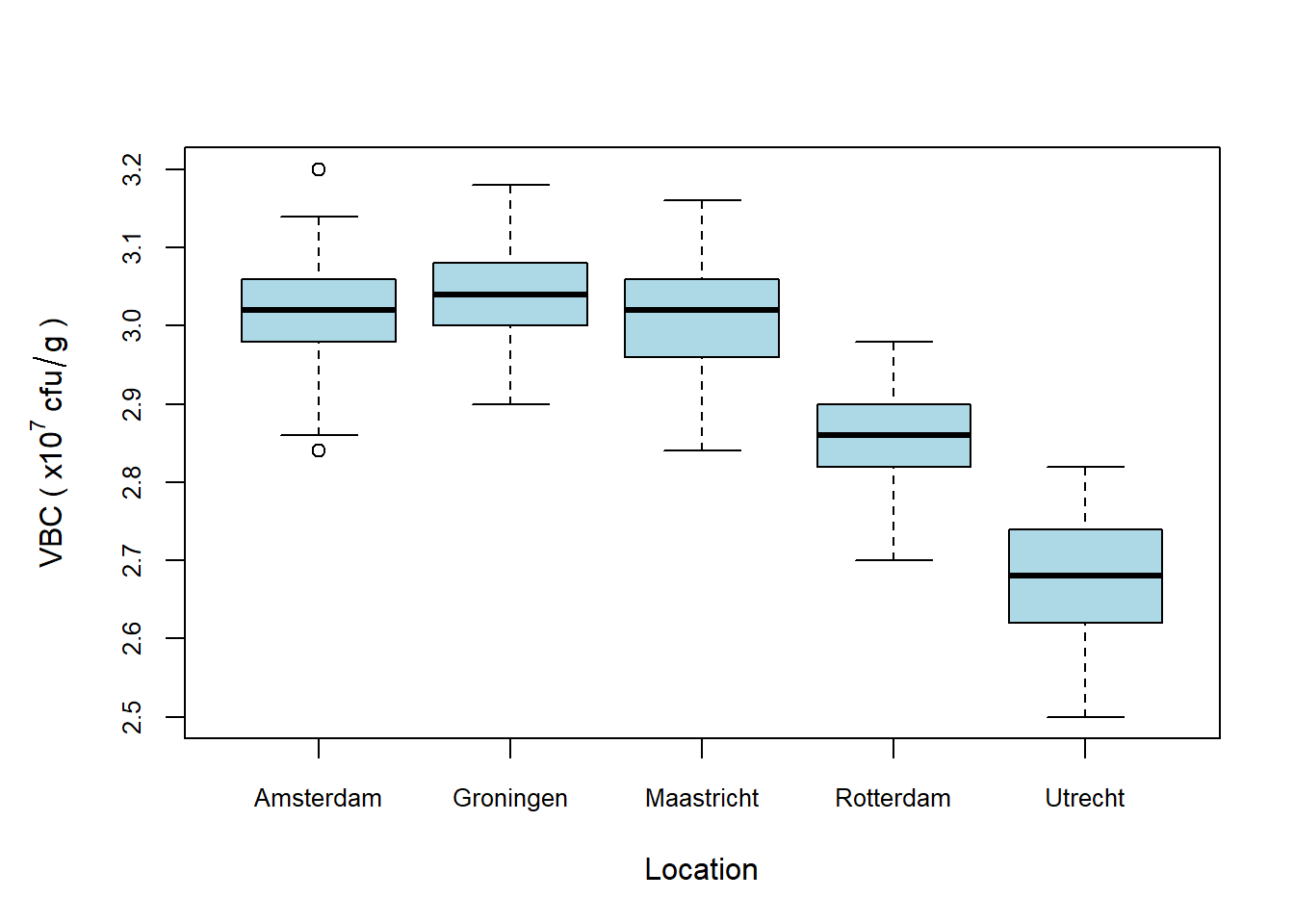
Figure 5.2: Viable bacterial counts (VBC) across five production sites. Rotterdam and Utrecht exhibit noticeably lower median VBC values compared to the other sites, indicating potential issues at these locations that may require further investigation.
You’ll notice that the first three lines of code are identical to the previous histogram example. Let’s break down the boxplot function.
- Adjust the margins to prevent y-axis label superscripts from being cut off
- Purpose: Annoyingly, if I use any label on the y-axis that has superscript elements then the default plot area in R isn’t quite big enough and they get cut off. The
par(mar = c(bottom, left, top, right))function adjusts the margins of the plot. By using a value of 5 for the second value (left margin), I’m ensuring that the y-axis is fully visible.
- Generate a grouped boxplot for VBC and group by location
boxplot(vbc ~ Location,
data = data,
main = "",
ylab = "Location",
xlab = expression(VBC ~ "(" ~ x10^7 ~ cfu/g ~ ")"),
col = "lightblue",
border = "black",
cex.axis = 0.8)- Purpose:
- Parameters:
vbc ~ Location: This specifies the formula for the boxplot. It creates boxplots of thevbccolumn data and groups it by theLocationfactor.data = data: Thedataargument specifies the data frame containing the variables in the above formula.cex.axis = 0.8: This adjusts the size of the boxplot label text, making it 80% of the default size. I like to do this as it improves readability.
Interpreting the Boxplot
For a reminder of the important features of a boxplot please refer to Anatomy of a Boxplot section in Chapter 2.
The key comparative insight gained from the boxplot is that there is a noticeable trend of lower VBC values at the Rotterdam and Utrecht sites. Whether this trend is statistically significant and the reasons behind the lower values at these two locations remain to be determined. However, if I were the CTO of MegaMush and you presented me with this boxplot, I would likely be making urgent phone calls to my colleagues at the Rotterdam and Utrecht sites to investigate the underlying causes of these discrepancies.
I hope you’ll agree that, unlike the histogram, which provides a general overview of data distribution, the boxplot offers a concise summary of central tendency and variability, making it easier to identify specific trends and outliers across different groups. This clarity is particularly valuable for decision-making, as it allows us to quickly pinpoint areas that require further exploration or intervention.
Ideas for Further Exploration
- Boxplot to show distribution of Temperature values grouped by Location.
- Boxplot to show distribution of Moisture values grouped by Location.
- Boxplots of VBC/Temperature/Moisture grouped by DayOfWeek
5.5 Scatterplot
The last thing on our todo list is a scatterplot. Remember, you’ve been asked to plot the relationship between Temperature and VBC. I also want to colour code the points by location. The code below shows how I’d do this with native R. Figure 5.3 shows your expected output.
# Your R code for generating a scatterplot.
# Load the data
data <- read.csv("compost_999.csv")
# Calculate the conversion factor
factor <- 2e5 / 1e7
# Calculate VBC column
data$vbc <- data$Viable.counts * factor
# Adjust the margins to prevent y-axis label from being cut off
par(mar = c(5, 5, 4, 2)) # Increase the second value for the left margin
# Define unique locations and corresponding colors
unique_locations <- sort(unique(data$Location))
colors <- rainbow(length(unique_locations), alpha=0.8) # Or use your predefined colors
# Create a scatterplot of VBC vs. temperature, colored by location
plot(data$Temperature, data$vbc,
main = "",
xlab = "Temperature (°C)",
ylab = expression(VBC ~ "(" ~ x10^7 ~ cfu/g ~ ")"),
pch = 19, # solid circles for points
cex = 1.5, # increase point size
col = colors[as.numeric(factor(data$Location, levels = unique_locations))]) # color by location with transparency
# Add a legend without a border
legend("bottomleft",
legend = unique_locations, # Use the unique locations
col = colors, # Ensure the correct color mapping
pch = 19, # same symbol as in the plot
bty = "n", # no box around the legend
pt.cex = 1.5) # match point size in the legend
# Add faint gridlines
grid(col = "gray90", lty = "dotted")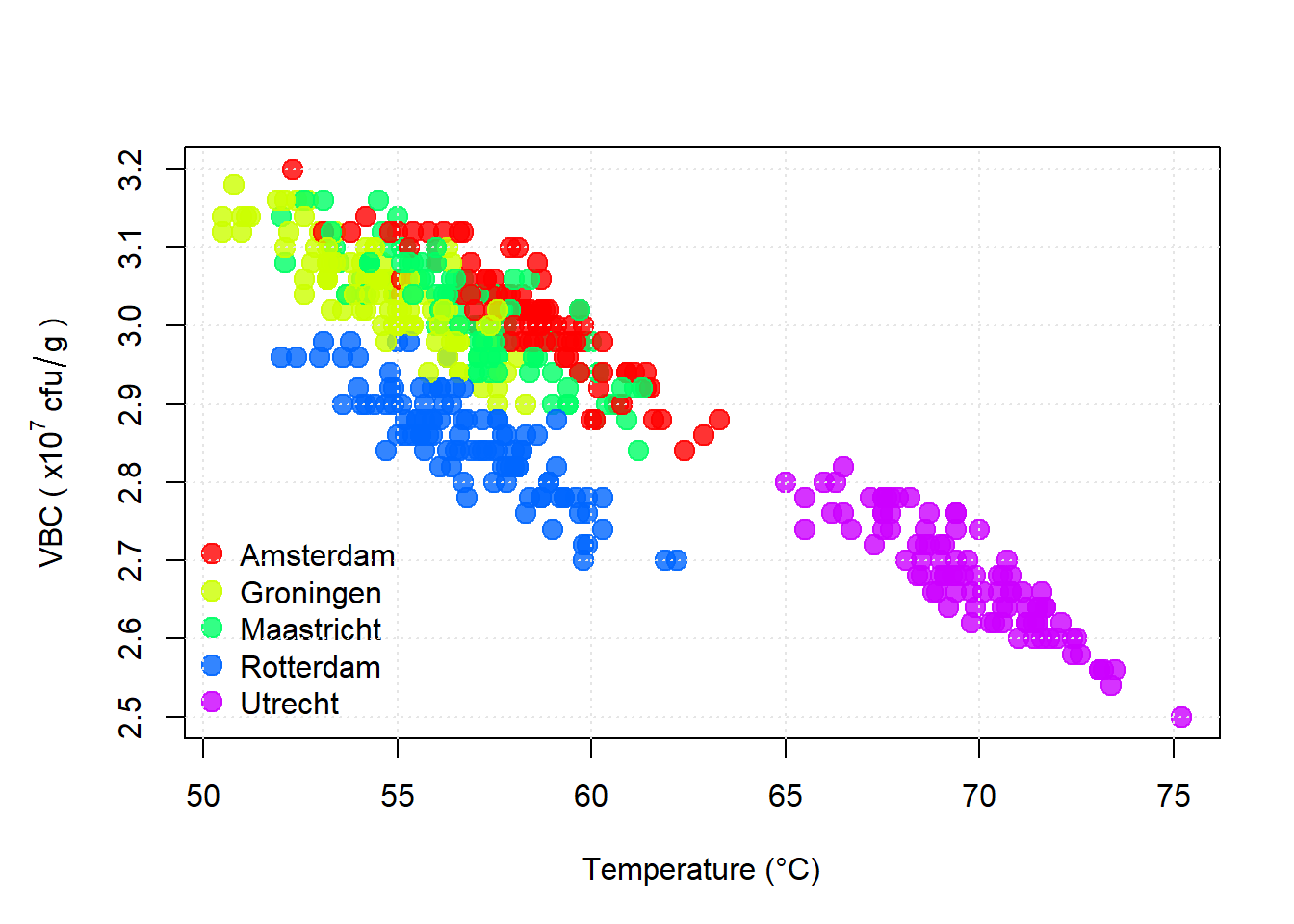
Figure 5.3: Viable bacterial counts (VBC) vs Temperature. A strong negative correlation is observed. Points are colour coded according to location (see key).
Create a new script file in your existing project, copy/paste the code above and then run as source. Hopefully, you should be able to recreate the scatterplot in your plots window. The first three lines of code are the same as for our boxplot example. Let’s break down the rest comment-by-comment:
- Define Unique Locations and Colors:
unique_locations <- sort(unique(data$Location))
colors <- rainbow(length(unique_locations), alpha = 0.8) # Or use your predefined colors- Purpose: to extract the unique production site locations from the data, sort them alphabetically, and assign each location a distinct colour for visualisation.
- Explanation:
unique(data$Location): finds all distinct location values (e.g., Amsterdam, Groningen, etc …).sort(): ensures the locations are listed in alphabetical order, which will also affect how they are represented in the legend.rainbow(length(unique), alpha =0.8): assigns a unique color to each location, creating a colour palette based on the number of unique locations. Thealpha = 0.8parameter makes the points slightly transparent.
- Create a Scatterplot of VBC vs. Temperature, coloured by Location:
plot(data$Temperature, data$vbc,
main = "",
xlab = "Temperature (°C)",
ylab = expression(VBC ~ "(" ~ x10^7 ~ cfu/g ~ ")"),
pch = 19, # solid circles for points
cex = 1.5, # increase point size
col = colors[as.numeric(factor(data$Location, levels = unique_locations))])- Purpose: To create a scatterplot that visualises the relationship between temperature and VBC, with points coloured according to the location of the data.
- Explanation:
plot(data$Temperature, data$vbc)creates a basic scatterplot of VBC values (on the y-axis) versus temperature (on the x-axis).xlabandylabprovide the axis labels.pch = 19specifies solid circular points for the scatterplot.cex = 1.5increases the size of points for better visibility.factor(data$Location, levels = unique_locations)ensures that the points are coloured by location in the correct, alphabetically sorted order. Theas.numeric()converts the factor levels into numeric indices to match with thecolorsvector.
- Add a Legend:
legend("bottomleft",
legend = unique_locations, # Use the unique locations
col = colors, # Ensure the correct color mapping
pch = 19, # same symbol as in the plot
bty = "n", # no box around the legend
pt.cex = 1.5)- Purpose: To add a legend to the plot that identifies the colours representing each location, ensuring the viewer can easily distinguish between the locations.
- Explanation:
bottom-leftdetermines the location of the legend on your plot.legend = unique_locationsensures the correct, alphabetically sorted location names are shown in the legend.col = colorsmaps the same colours used in the plot to the corresponding locations in the legend.pch = 19matches the symbol style of the points in the plot with those in the legend.bty = "n"removes the default box around the legend to keep the visual clean.pt.cex = 1.5adjusts the size of the symbols in the legend to match this size of the points in the scatterplot.
- Add Gridlines:
- Purpose: Pretty self-explanitory this one I think. I like to have gridlines on my scatterplot as a guide to for the eye, but I leave it as optional for you.
Interpreting the Scatterplot
There’s lots of information radiating out of this plot. Let’s interpret what we’re seeing:
- Overall Trend: A clear negative correlation between temperature and VBC.
- Utrecht (purple): Highest temperatures (65 - 75oC) and lowest VBC values.
- Rotterdam (blue): Slightly lower VBC values for the same range of temperatures as Amsterdam and Groningen. This suggests that there may be another factor affecting VBC at Rotterdam.
After seeing this scatterplot I’d be advising the Utrecht site that they were “running hot!” and that they need to cool their compost by about 15oC in order to bring their VBCs back in line with the other sites.
Ideas for Further Exploration
- Scatterplot of VBC vs Moisture
- Scatterplot of Moisture vs Temperature.
5.6 Just for Fun
In this last sub-section I just wanted to show you something really cool. The code below uses the fantastlic Plotly library to generate a 3D scatterplot so that I can see the impact of both Temperature and Moisture on VBC at the same time. Plotly is not included in your RStudio by default, you’ll need to install it first. Go to your Console window and type:
Hit enter and then wait while the Plotly package is installed. Then start a new script file, copy/past the script below and run it as source.
# Include the plotly library in your environment
library(plotly)
# Load the data
data <- read.csv("compost_999.csv")
# Calculate the conversion factor for cfu per gram
factor <- 2e5 / 1e7
data$vbc <- data$Viable.counts * factor
# Create an interactive 3D scatterplot
fig <- plot_ly(data = data,
x = ~Temperature,
y = ~Moisture,
z = ~vbc,
color = ~Location,
colors = "Set2", # Choose a color palette
type = 'scatter3d',
mode = 'markers',
marker = list(size = 5)) %>%
layout(scene = list(xaxis = list(title = 'Temperature (°C)'),
zaxis = list(title = 'VBC'),
yaxis = list(title = 'Moisture (%)')))
# Show the figure
figFigure 5.4: A 3D scatterplot showing Viable bacterial counts (VBC) vs both Temperature AND Moisture using the fantastic Plotly package.
How cool is that? You have an interactive, 3D graph that you can fiddle with and investigate the relationship between VBC, Temperature and Moisture.
5.7 Complete your Weekly Assignments
In the BIOS103 Canvas course you will find this week’s summative QS assignments. There is no formative assignment this week! You should aim to complete the summative assignment before the end of the online workshop that corresponds to this section’s content.
- You can attempt the summative assignment only once. This assignment will contribute to your overall score for this course. Failure to complete a summative test before the stated deadline will result in a zero score. You will not receive immediate feedback after submitting summative assignments. Typically, your scores will be posted within 7 days.
In ALL cases, when you click the button to “begin” a test you will have two hours to complete and submit the questions. If the test times out it will automatically submit.